*pictures at end*
- explain the structure of DNA – use the terms nucleotides, antiparallel strands and a complementary base pairing.
- DNA (Deoxyribonucleic acid) is shaped in a structure called a double helix, which looks like a twisted ladder. The ladder structure is made up of nucleotides that form the ladder shape. The DNA backbone is made up of alternating phosphates and deoxyribose sugars connected through a covalent bond, these backbones run antiparallel to each other with a 5 end and a 3 end that always run opposite. the ‘rungs’ of the ladder are made up of the four bases which are bonded through hydrogen bonds. Adenine and Thymine always bond together and Cytosine and Guanine always bond together, this is known as a complementary base pairing. A base (adenine, thymine, cytosine, guanine) bonded to deoxyribose, bonded to phosphate is a nucleotide, which is the monomer of nucleic acid. When many nucleotides bond together, including complimentary base pairings, and antiparallel strands the result is a strand of DNA. (1a, 1b)
- When does DNA replication occur?
- DNA replication occurs before cell division during the S stage of interphase.
- Name and describe the 3 steps involved in DNA replication. Why does the process occur differently on the “lagging” strands?
- unwinding and unzipping: a DNA helicase unwinds the double-helix of the DNA and breaks the hydrogen bond between the complementary base pairings. (3a)
- complementary base pairing: the DNA polymerase scans the separated DNA strands and attaches complementary base pairs to each of the nucleotides joining of adjacent nucleotides: due to the way the DNA polymerase works (spaces are left between nucleotides on the backbone because of where it starts and ends), the DNA ligase but join adjacent nucleotides to complete the structure. (3b)
- Since the DNA polymerase can only read starting at the 3 directions, the complementary strand will be built in segments moving backward starting at the ‘replication fork’ (where the strands diverge). (3c)
- Today’s modeling activity was intended to show the steps involved in DNA replication. what did you do to model the complementary base pairing and joining of adjacent nucleotides steps? In what way was this activity well suited to showing this process? In what way was it inaccurate?
- to model the complementary base pairing we showed how showed how the DNA helicase cut the DNA and then how the polymerase was followed by paired nucleotides. For the joining of adjacent nucleotides, we showed (as mentioned above) how the DNA polymerase was followed by bonded nucleotide, but we took the picture with the polymerase in the middle with some of the bases paired at the start and at the end (how it happens with the lagging strand), we then used the heart-shaped (for this example) ligas to show how it joined the adjacent nucleotides.
- we skipped the part of the DNA unwinding, which is essential to DNA replication. (inaccurate)
- we were able to model the different steps and use a helicase, polymerase and ligase that, though they didn’t look like their real-life shape, helped us understand the roll they play (ex. ligas shaped as a heart to show how it bonds adjacent nucleotides). The large scale helped to see how the process works (4a)
- 5. how is mRNA different than DNA?
- DNA is very large and important, therefore it cannot leave the nucleus. mRNA is a copy of DNA’s instructions which are taken from a single gene (about 1000 nucleotides long) which may leave the nucleus through a nuclear pore, move to the cytoplasm and it allows the ribosomes to pull together amino acids to form a protein.
- DNA holds all the genetic information, while mRNA only holds one gene
- DNA’s sugar is Deoxyribose while mRNA’s sugar is Ribose
- DNA has the base thymine while mRNA has the base uracil.
- 6.Describe the process of transcription
- the DNA unwinds and unzips, the complementary bases are paired Adenine & Uracil, Guanine & Cytosine. Then the mRNA separates from the DNA, it gets modified, then it exits the nucleus.
- 7. How did today’s activity do a good job of modeling the process of RNA transcription? In what ways was our model inaccurate?
- the large scale helped to see how the process works, we were able to model the differences with the DNA and mRNA and using different colours helped us see the different sugars that each nucleic acid has.
- (inaccurate) we skipped some steps such as how exactly the mRNA separates from the sense strand, the modification of the mRNA, and what happens to the other DNA back-bone while the sense strand is being used for the production of mRNA.
- 8. describe the process of translation: initiation, elongation, and termination
- translation: when the DNA gets transcribed into RNA, through complementary base pairings. (8a)
- elongation: amino acids gets stacked to create a protein chain(8b-e)
- termination: the chain of amino acids stops once it reads a codon that has no pair. (8f)
- 9. how did today’s activity do a good job of modeling the process of translation? in what ways was our model inaccurate?
- it helped us to see the roll that DNA, mRNA, tRNA, and amino acids play in the production of proteins and how the amino acids get stacked into their order.
- we did a condensed version of translations and skipped some steps (skipped the mRNA leaving the nucleus
- the shapes were not accurate
Images:
DNA:
1a)
1b)
3a)
3b) 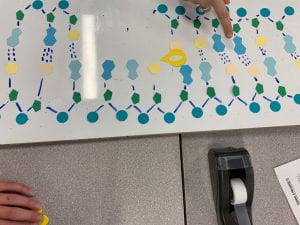
3c) finished
finished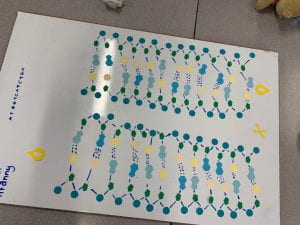
mRNA
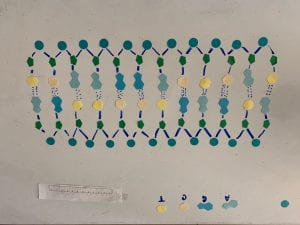
^Original DNA

^ DNA un unwound and unzipped
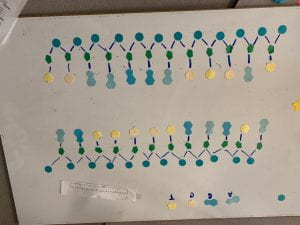
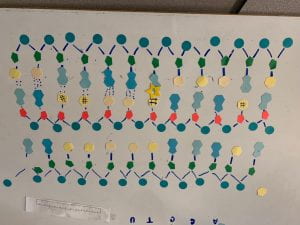
^ complementary base parings (only happens on the sense strand)

^ mRNA beaks off from DNA (moves on to the next steps)
Protein synthesis
8a)
8b) 8c)
8c)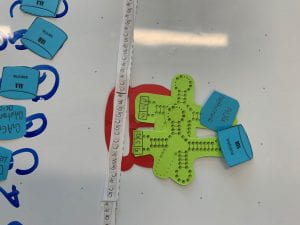
8d) 8e)
8e)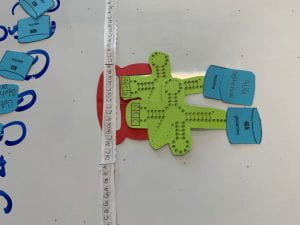
8f)
