1. Explain the structure of DNA – use the terms nucleotides, antiparallel strands, and complimentary base pairing.
DNA (Deoxyribonucleic acid) is a large polymer consisting of 4 nucleotides, each made up of a phosphate group, a sugar group (deoxyribose), and a nitrogenous base. Two of which are double ringed (Purines) and two which are single ringed (Pyrimidine).
Purines -> Adenine & Guanine | Pyrimidines -> Thymine & Cytosine
The two backbones of DNA are formed through the bonding of the sugar-phosphate pieces of adjacent nucleotides. The nucleotide bases face “into” the ladder and are formed through H-bonds that lie between the base pairs. The bases attach together in a special way, always with the same partner, known as complimentary base pairing; Adenine -> Thymine & Guanine -> Cytosine. The base pairs form two long strands that spiral, creating a double helix. The two DNA strands are antiparallel, they lie parallel to one another yet are read in opposite directions; and complimentary, this is when the strands give the same message.
2. How does this activity help model the structure of DNA? What changes could we make to improve the accuracy of this model? Be detailed and constructive.
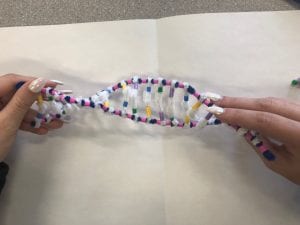
The activity helps model the structure of DNA through the use of pipecleaners and beads to show its different parts. Pipecleaners are used to show both the two backbones as well as for the H-bonds that are formed; beads are used to represent the 4 complimentary bases, as well as the number of rings each base contains. We used two beads for the purines to show their double ring and, one bead for the pyrimidines to show their single ring. The different colours used for the bases help make a clear visual on complementary base pairing. The model we made also accurately demonstrates how the strands are antiparallel to one another. Some changes to make this activity more accurate would be to have accurate measurements included in order to show the spacing between everything on the DNA strands. Another is that the activity doesn’t accurately demonstrate the number of hydrogen bonds that are formed between the base pairs, there could be an increase in the number of white pipecleaners used to show the two bonds formed between adenine and thymine and the three bonds formed between guanine and cytosine.
3. When does DNA replication occur?
DNA replication occurs prior to cell division. It is a semi-conservative process as each new molecule must contain one backbone from the original DNA strand, allowing each new molecule to have the same instructions to make new proteins.
4. Name and describe the 3 steps involved in DNA replication. Why does the process occur differently on the “leading” and “lagging” strands.
DNA replication occurs in 3 stages: Unwinding, complimentary base pairing, and joining.
Step 1: Unwinding – The DNA’s double helix structure begins to unwind, the enzyme known as helicase “breaks” the hydrogen bonds that are holding the complimentary base pairs together. The separation of two single strands of DNA creates a shape referred to as the replication fork.
Step 2: Complimentary base pairing – Nucleotides present in the nucleus move and form a H-bond with “partners” on the template strands of DNA. This process is facilitated by DNA polymerase. One strand is in the 3′ to 5′ direction (towards the replication fork), this is the leading strand. The other strand is in the 5′ to 3′ direction (away from the replication fork), this is the lagging strand. Replication that occurs on the leading strand is continuous. A primer binds to the end of the leading strand and acts as a starting point for DNA synthesis. DNA polymerase binds to the leading strand and ‘walks’ along it, adding new complimentary bases to the strand in a 5′ to 3′ direction. Replication that occurs on the lagging strand is discontinuous. A number of RNA primers bind to the lagging strand at various points. Parts of DNA (Okazaki fragments), are then added to the lagging strand in the 5′ to 3′ direction. The reason the lagging strands replication is discontinuous is due to the fact that the Okazaki fragments need to join up later.
Step 3: Joining – Covalent bonds form between nucleotides on the new strand. The leading strand will remain continuous as the DNA “unzips”. On the lagging strand, fragments will begin to form as the DNA “unzips”; the fragments will be glued together by DNA ligase. As a result two daughter strands are formed.
5. The model today wasn’t a great fit for the process we were exploring. What did you do to model the complementary base pairing and joining of adjacent nucleotides steps of DNA replication. In what ways was this activity well suited to showing this process? In what ways was it inaccurate?
To model complimentary base pairing we paired some of the ‘free’ nucleotides to the complimentary base on the template strand. To show the joining of adjacent nucleotides we attached the newly based pairs to one of the backbones.
This activity gave a good visual on how the leading and lagging strands are read. It also demonstrated how the bases were attached to the leading strand. Some of the things that made this activity inaccurate was the use of the candies. We only placed the candies in the areas where we thought they would be placed and function, we didn’t get to see how they interact and work. We also knew where to place DNA polymerase yet, it didn’t really help show its function and how it interacts with the lagging strand. It was easy to understand how the DNA polymerase interacted with the leading strand but was harder to grasp and show for the lagging strand.