DNA Model
1. Explain the structure of DNA – use the terms nucleotides, antiparallel strands, and complimentary based pairing.
Deoxyribonucleic Acid also known as DNA is the makeup of chromosomes and genes, located in the nucleus of cells. It is a double helix structured polymer, made up of 6 smaller molecules known as the nucleotide monomers. Each nucleotide consists of a 5-carbon sugar: deoxyribose, phosphate and a nitrogen base. The two back-bones of DNA are made up of the sugar-phosphate portion of the nucleotide. The nitrogen bases attached face inwards to make up the rungs of the ladder shape that is made of DNA, when flattened. There are two categories of nitrogen bases in DNA: purines (Adenine {A}, Guanine{G}) and pyrimidines (Thymine {T}, Cytosine {C}). Purines have a double-ring base and Pyrimidines have only a single ring base. A only bonds with T and G only bonds with C, they are bonded by hydrogen bonds. G and C are held together by 3 hydrogen bonds making it a stronger bond then A and T, held together by only 2 hydrogen bonds. This is known as complimentary based pairing and because of this the strands are complimentary to each other, you can read either strand for the same info because you know that A always has T on the other side and C always have G on the other side, and vice versa. Finally, the two strands are connected antiparallel, the leading strand goes from 3′ to 5′ and the lagging strand from 5′ to 3′. The 5′ end has a phosphate group and the 3′ end has a hydroxyl. The strands can only be read from 3′ to 5′ so they are ready in opposite directions: ⇅.
2. How does this activity help model the structure of DNA? What changes could we make to improve the accuracy of this model? be detailed and constructive.
In this activity we were able to demonstrate the double helix structure and the double backbone that DNA possesses. In order to represent the nucleotides we used beads and specific colours for each molecule. We demonstrated complimentary based pairing by showing: Adenine (yellow bead) bonding with Thymine (Blue bead) and Guanine (Purple bead) bonding with Cytosine (Green bead). We represented Adenine and Guanine’s double-ring base using two beads of those colours instead of one. We represented the phosphate-sugar group with pink beads.We were even able to represent the difference between 3′ and 5′ by distinguishing the 5′ strand with a sugar-phosphate molecule on top (pink bead).
To improve the accuracy:
- We could have used two beads of different shapes to represent the phosphate-sugar group to distinguish between the 5 carbon sugar and the phosphate circle.
- We could have used another shape to demonstrate the hydroxyl (-OH) found at the 3′ end of the backbone.
- We missed a phosphate-sugar group at the end of our leading strand.
- We did not distinguish the difference between the 3 hydrogen bonds holding Guanine and Cytosine versus the 2 hydrogen bonds holding Adenine and Thymine.
DNA Model Replication
- When does DNA replication occur?
DNA replication occurs before cell division – the growing, repairment and replacement of cells. It occurs during the interphase of mitosis. The functions of DNA include the controlling of tasks in the cell, protein building and controlling the energy usage of the cell.
2. Name and describe the 3 steps involved in in DNA replication. Why does the process occur differently on the “leading” and “lagging” strands.
DNA replication is semiconservative. This means that one strand is conserved (the parent strand) while the other strand is formed through replication (daughter strand) The three steps in semiconservative DNA replication are:
a. Unwinding and Unzipping
Unwinding and Unzipping is the process of the DNA strands separating from one another forming a template for the new strand – the complementary back bone. The double helix shape that DNA holds, unwinds flattening into a ladder shape.
The hydrogen bonds between the left and right side bases are broken up by the zipper like enzyme known as the helicase molecule. The open part of the DNA is a template for the new strand.
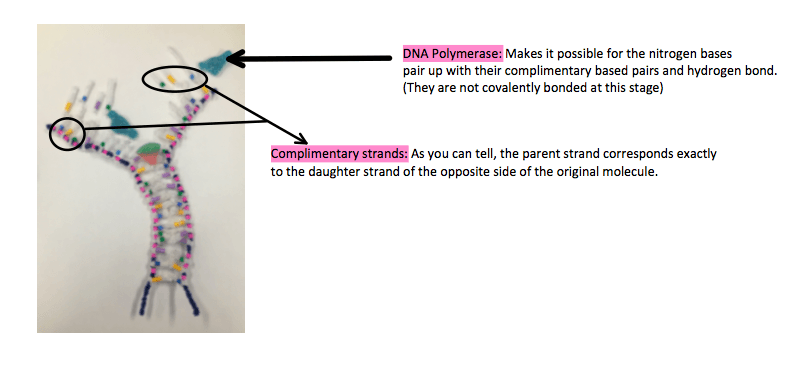
b. Complimentary Based Pairing
The original pairings are now separate and the new nitrogen bases pair up with their complimentary pairings. It’s the same pairing as before. C bonds with G (3 hydrogen bonds) and A bonds with T (2 hydrogen bonds) A new complimentary backbone is beginning to form. DNA polymerase – the enzyme that joins together makes this step possible. All molecules are separate and do not join together at this stage.
c. Joining
The nucleotides are now joined together by covalent bonds. The strands must be built from 5′ to 3′ which means that they must be read from 3′ to 5′ because they are antiparallel.
The leading strand is 3′ to 5′ therefore it is built and read by DNA polymerase in a continuous manner from the 3′ end to the 5′ end.
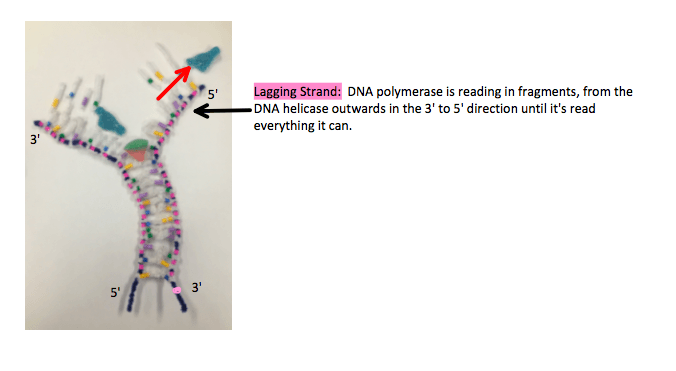
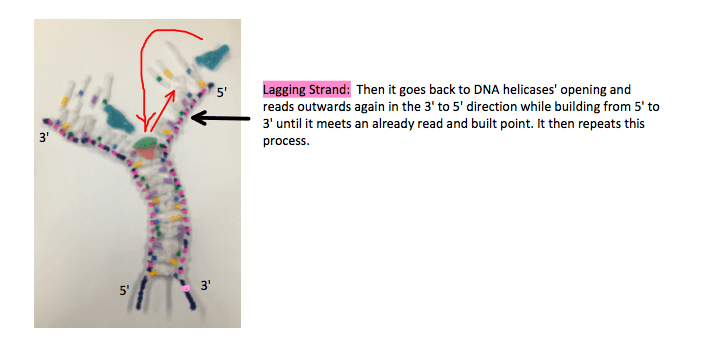
The lagging strand however, is 5′ to 3′ and has to be read in the 3′ to 5′ direction, which is backwards. DNA polymerase starts where the DNA helicase is unzipping and reads the strand backwards in fragments, when it reaches the end it returns to the newest opening and reads in the 3′ 5o 5′ direction while building a 5′ to 3′ antiparallel strand to where it has already read, and repeats the process.
It’s discontinuous and creates fragments, as it has to keep going back to the new openings. The enzyme, DNA ligase glues these fragmented strands together.
Once the process of semiconservative replication is finished, there are two DNA molecules. Each one contains one parent and one daughter strand. Both molecules are identical to the original. Because of complimentary based pairings. Because each nitrogen base only has one pairing option, a limitless amount of copies can be made from only one DNA strand, thus promoting the continuity of life.
3. The model today wasn’t a great fit for the process we were exploring. What did you do to model the complementary based pairing and joining of adjacent nucleotides steps of DNA replication. In what ways was this activity well suited to showing this process? In what ways it inaccurate?
- We were able to demonstrate the unwinding and unzipping process.
- We were able to demonstrate the way that the nitrogen bases pair up in the same complimentary based pairs every time.
- It was difficult to demonstrate how during the process of complimentary based pairing the nucleotides were H-bonded but not covalently bonded, we didn’t really demonstrate that well, and just photographed them lying freely next to their pairs.
- We also scratched the surface of the direction that the DNA polymerase travels in very briefly but could have done a better job visually demonstrating, in more steps how the lagging strand works. We didn’t demonstrate okazaki fragments using pipe cleaners and the process of the covalent bonding of the back-bones because we didn’t have cut sections of the blue pipe cleaner, in lieu of this I drew it in the photo I embedded, when explaining DNA ligase.