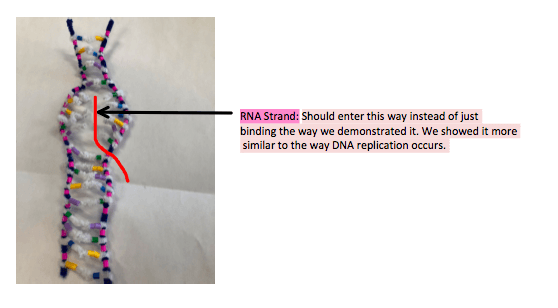RNA Transcription Model
- How is mRNA different then DNA
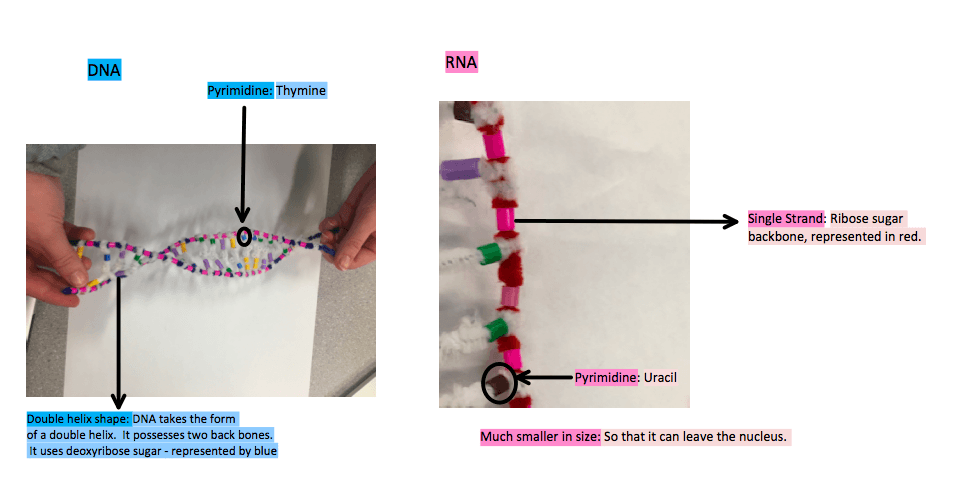
mRNA differs from DNA in the sense that it uses the ribose sugar in lieu of deoxyribose. It is shorter then DNA which allows it to exit from the nuclear pore – something that DNA is not capable of. It has one strand instead of two, making it a single backbone. It does not have an double helix shape. mRNA substitutes the pyrimidine uracil for the pyrimidine thymine, meaning that all the Adenines on DNA are paired with Uracil on RNA.
- Describe the process of transcription
This whole process is essential because DNA carries the information for protein synthesis but is too large and vital to the nucleus to exit. Protein synthesis must occur in the cytoplasm of the cell where the ribosomes, or protein factories are located. mRNA is derived form the process of transcription in order to be able to exit the nuclear pores with DNA’s information and produce codons to be read by the ribosomes later in translation for the construction of proteins.
- Unwinding and unzipping of DNA
DNA helix shape unwinds. The unzipping of DNA only happens at the desired location of the gene for specific information. Exposing instructions to build the gene that is running low only. The whole 85 million long nucleotide strand does not unzip. The sense strand is the strand that RNA has it’s information transcribed from. The sense strand is the strand that carries the information for protein synthesis. The nonsense strand is useless in this process as it would spit out non readable info if transcribed, it bonds as a compliment to the sense strand on DNA.
2. Complimentary based pairing with DNA
Hydrogen bonds are formed between the complimentary based pairs. RNA does not posses the pyrimidine thymine and instead the pyrimidine uracil is bonded to adenine. Guanine and Cytosine remain pairs. The RNA strand (in red) is base pairing with the nitrogen bases on the sense strand (in blue). This is essential so that the information is copied precisely.
3.The RNA polymerase Enzyme, represented by a fuzzy peach in this picture facilitates the bonding between the nearby nucleotides. There is a sugar & phosphate covalent bond.
4. Separation from DNA
RNA is separated from DNA, and DNA re-zips and returns to it’s original double helix shape.
Its’ the messenger RNA (mRNA) that is now developed by the process of replication of the information the gene on DNA possessed. It is checked and refined to ensure that the introns (unnecessary sections) are removed as well as to ensure that it is capped at the ends. It then can exit through the nuclear pore.
- How did today’s activity do a good job of modelling the process of RNA transcription? In what ways was our model inaccurate.
Accuracies:
- We modelled only one strand for RNA and two strands in a helix shape for DNA.
- We used a red strand to distinguish ribose sugar and blue for deoxyribose sugar.
- We used a brown bead for Uracil the pyrimidine that binds with Adenine in RNA and a Blue beed for Thymine the pyrimidine that binds with Adenine in DNA.
- We modelled RNA polymerase using a fuzzy peach showing how it bonds the nucleotides in a row.
Inaccuracies:
- We demonstrated the transcription on our entire model as it was only 18 nucleotides long however generally the RNA strand should open up, as we demonstrated in the unwinding and unzipping process and then the RNA strand should enter like so:
- It’s inaccurate how the sense strand and the RNA strand were connected with no sign of the complimentary strand as the above photo shows that the RNA strand should enter inside the double bond of DNA and even thought it has unzipped the RNA strand should not completely replaced the complimentary strand, as the model showed in step 2:
- We did not demonstrate the process of the mRNA being refined before modelling translation down below.
Protein Synthesis Model
- Describe the process of translation: initiation, elongation and termination.
Prior to Translation, the process of transcription (as shown above) must occur in order for mRNA to be derived from the DNA sense strand. Here is a short summary:
- Initiation
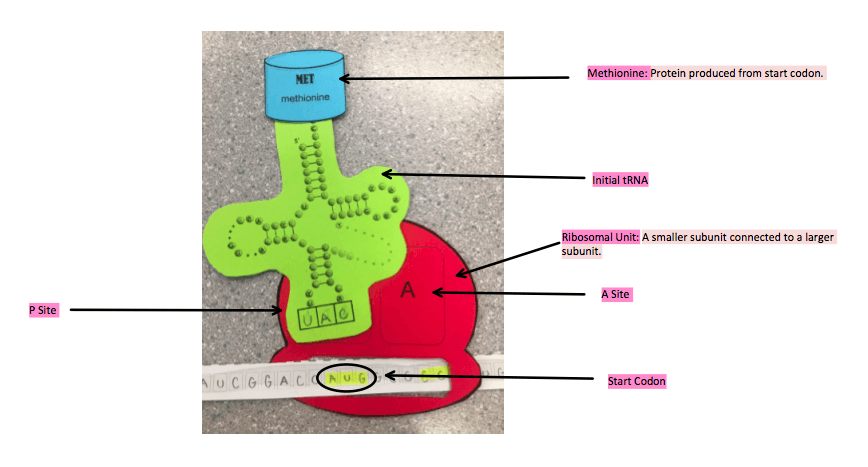
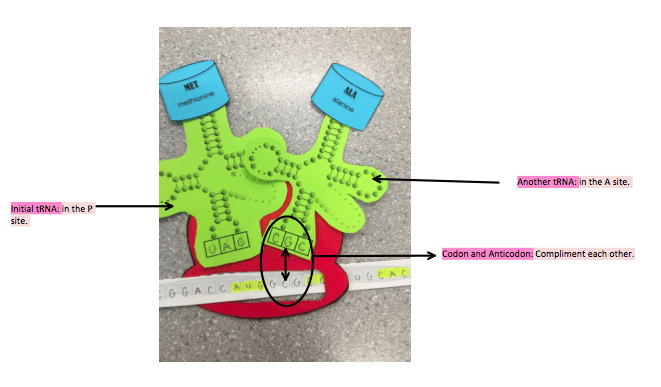
The mRNA released from the nuclear pores and connected by transcription bonds with a small ribosome subunit in the cytoplasm. The small and large ribosome subunits then bond together. Ribosome starts to read the mRNA that has bonded to it and waits for the start codon AUG. mRNA start codon is AUG, codons produced by mRNA are complimented by anticodons produced by tRNA. There are 64 3 letter codons and from these 64: 20 amino acids can be formed. There are two binding sites in the ribosome, one is the P site and one is the A site. This means that a ribosome can contain two tRNA’s at a time. The initial tRNA binds to the P site, and it contains one amino acid Methionine the amino acid formed by the start codon AUG.
2. Elongation
Another tRNA binds in the A site:
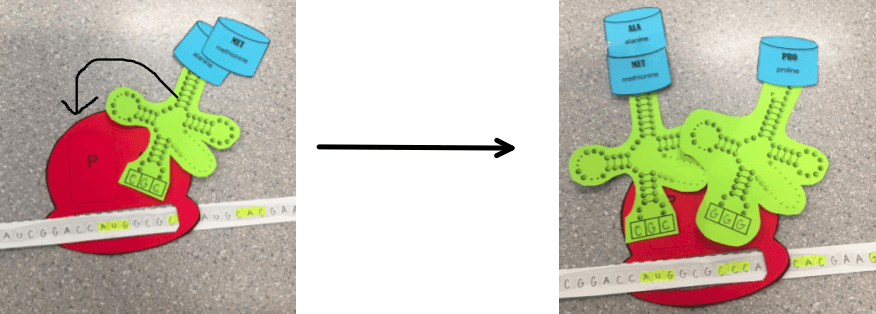
The amino acid from the initial tRNA moves to bond with the amino acid in the new tRNA in the A site. The initial tRNA is released.
This opens up a space and the remaining tRNA is moved to the P site. A new tRNA binds in the A site.
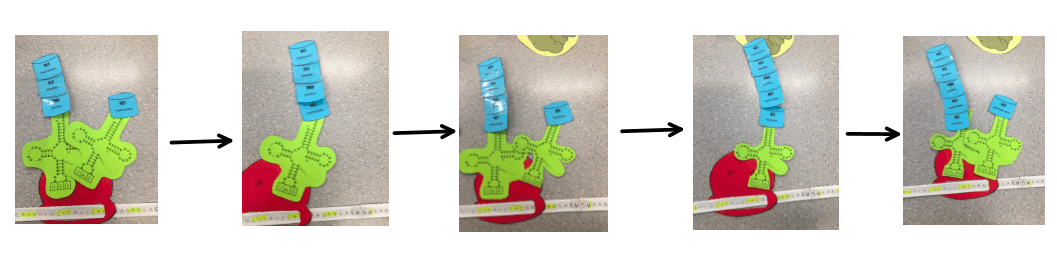
The process continues as the amino acids from the tRNA In the P site are moved to the A site so that the tRNA in the P site can be released. Freeing up a space so that the tRNA in the A site can move over to the P site, making room for the a new tRNA.
3. Termination
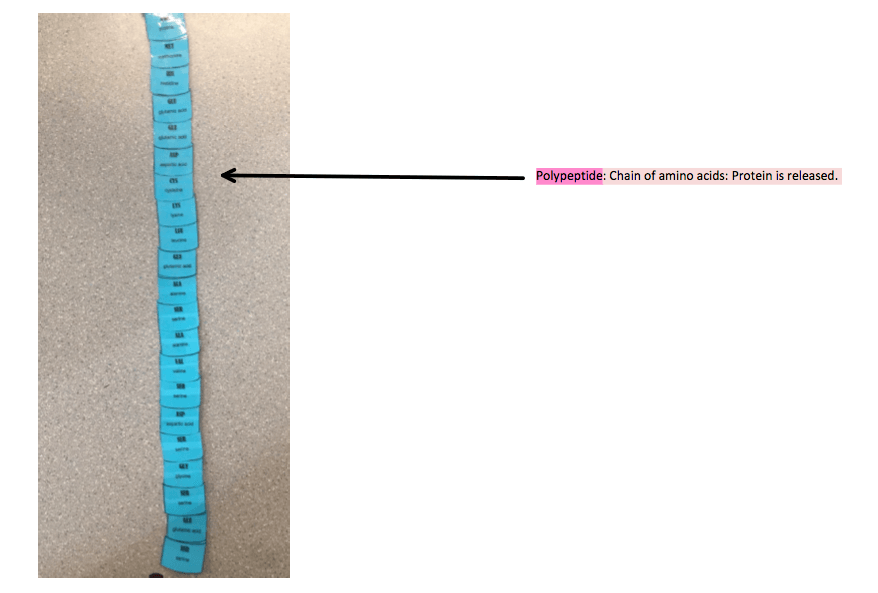
A stop codon is read on the mRNA – UAG, UGA or UAA. There is no amino acid that is produced from any of these stop codons which means that the process is terminated. A release factor binds in the A site, in lieu of a new tRNA, causing the breakage of the bond between the tRNA and the P site (the final tRNA). The subunits of the ribosome disconnect and the polypeptide chain of amino acids – the protein, is released.
- How did today’s activity do a good job of modelling the process of translation? In what ways was our model inaccurate.
Accuracies:
- The start codon was modelled, as well as the anticodon that complimented it, as shown in the initiation process above. Methionine was accurately the first protein.
- The two sites (A and P) were shown as well as the process of the new tRNA bind in the A site and the proteins transfer from the older tRNA in the P site with the newest protein in the A site.
- The process of elongation was modelled nicely.
Inaccuracies:
- We did not show the initial smaller ribosome subunit bonding with only the initial tRNA. We showed the initial tRNA bonding with the entire ribosomal unit when in the textbook it explains that the initial tRNA bonds with only the smaller ribosomal unit and then bonding afterwards with the larger ribosomal subunit for completion of the ribosome.
- We did not model the stop codon UAG, UGA or UAA.
- We did not model a release factor bonding in the A site in order for the polypeptide to dissociate and get released.
- We did not model the separation of the ribosomal subunits just as we did not model when they bind together. The separation occurs with the release of the polypeptide.
- Finally, we could have modelled it better, by showing the release of the tRNA each time a little more clearer then simply having it disappear. We could have shown it outside the ribosomal unit.