
(Inserted above is my group’s datasheet for the agar cube lab. All our information for our lab is recorded above.)
- In terms of maximizing diffusion, what was the most effective size cube that you tested?
The most effective size cube that we tested was the 1cm cube. This cube was the best at diffusion and the percent diffusion was 100% compared to the 2cm cube which has a percent diffusion of 81.25% and the 3cm cube with 70.37%.
 ( This is a photo of the agar cubes cut in half to see the diffusion after a 10 minute period.).
( This is a photo of the agar cubes cut in half to see the diffusion after a 10 minute period.).
2. Why was that size most effective at maximizing diffusion? What are the important factors that affect how materials diffuse into cells or tissues?
The 1cm cube was most effective at maximizing diffusion as it has the highest surface area ratio and the lowest volume ratio (6:1). The 6 represents the surface area and the 1 represents the volume. For successful diffusion, the ratio should have a high surface area and a low volume. The important factors that affect how materials diffuse into cells or tissues that we touched on in this lab is the surface area to volume ratio; however, other factors may include temperature, concentration, density, and pressure.
3. If a large surface area is helpful to cells, why do cells not grow to be very large.
A large surface area is helpful to cells, through this lab we could see that the more surface area cells and lower volume in the ratio are ideal for the most effective diffusion. Cells do not grow to be very large because if they increase in size, they increase volume which in turn will increase the SA: V ratio. This means that in reality, diffusion becomes harder and less effective if cells were to grow very large. Cells need to be small because they rely on diffusion for getting substances into and out of their cells.
4. You have three cubes, A, B, and C. They have surface to volume ratios of 3:1, 5:2, and 4:1 respectively. Which of these cubes is going to be the most effective at maximizing diffusion, how do you know this?
Cube C (4:1) will be most effective at maximizing diffusion as it has the highest Surface Area in the SA: V ratio.
5. How does your body adapt surface area-to-volume ratios to help exchange gases?
Your body can adapt its ratios in a number of ways such as: folding the surface of the object/ cell membrane, being shaped differently to perform specific functions, or/and cells divide when they get to bid to restore/have a larger SAV ratio. Producing a high SAV ratio is very important as this allows us to have a good gas exchange rate; however, if our surface are drops, so does our rate of gas exchange.
6. Why can’t certain cells, like bacteria, get to be the size of a small fish?
Bacteria is unicellular and must be able to sustain itself, as opposed to a small fish that is multi-cellular and is composed of many different cells. Bacteria need to be able to diffuse efficiently as if it can not it will die. If the bacteria became the size of a small fish the surface area would grow but so would the volume and as a result, the diffusion is not as effective and the bacteria will die.
7. What are the advantages of large organisms being multicellular?
By being multicellular organisms, plants and animals have overcome the problems of small cell sizes. Each cell has a large SAVratio which is necessary as there are a lot of functions needed to be performed with their specialized systems and cells. These features include gas exchange organs (lungs) and circulatory system (blood) to speed up and aid the movement of materials into and out of the organism. Furthermore, by being multicellular the cells in large organisms can vary in size if necessary.
- What is the difference between DNA and mRNA.
While DNA and mRNA both play necessary roles in the human bodies, they both differ from one another. DNA is double-stranded and shaped like a double-helix, whereas, mRNA is single-stranded and appears as a single spiral. Furthermore, DNA has the sugar deoxyribose sugar and mRNA has the ribose sugar. Not only do they differ in sugars, but they also differ in nitrogen bases. DNA has the base Thymine, Cytosine, Adenine, and Guanine, whereas, mRNA has the base Uracil, Cytosine, Adenine, and Guanine. Lastly, mRNA carries the genetic information from the DNA to a ribosome, in contrast to DNA which carries genetic information.
2. Describe the process of transcription.
Transcription is the process by which the information in a strand of DNA is copied into a new molecule of messenger RNA (mRNA). This transcription process occurs in multiple steps, all of which occur due to the enzyme, RNA polymerase. Essentially, Transcription is how the message is prepared to be sent off and received.
The first step of Transcription is Unwinding and Unzipping. A specific section of the DNA molecule unwinds, revealing a set of bases (a gene). The H-bonds between nitrogen bases in this specific section have been broken to allow for RNA polymerase to come and continue to the next step of the Transcription process.
 (This image is DNA unwound from its original double-helix shape. It is now resembling a ladder. Furthermore, this image does not include the unzipping stage of Unwinding and Unzipping.)
(This image is DNA unwound from its original double-helix shape. It is now resembling a ladder. Furthermore, this image does not include the unzipping stage of Unwinding and Unzipping.)
Step two in the process of Transcription is Complimentary Base Pairing with DNA. RNA polymerase reads the unwound DNA strand and builds the mRNA molecule, using complementary base pairs. This is only along one strand, the sense strand. This is the strand that carries the correct instructions for building the protein, unlike the nonsense strand which if transcribed would not create a protein. Complementary base pairs are made between nitrogen bases (these are hydrogen bonds). This is the same concept as in DNA molecules, however, since RNA (represented by a red pipe cleaner) does not contain thymine as instead of thymine it contains Uracil. The Uracil will pair with the Adenine and the Guanine and Cytosine still pair together. The RNA polymerase also creates the forming of covalent bonds between adjacent nucleotides to build the RNA backbone (phosphate and ribose sugar).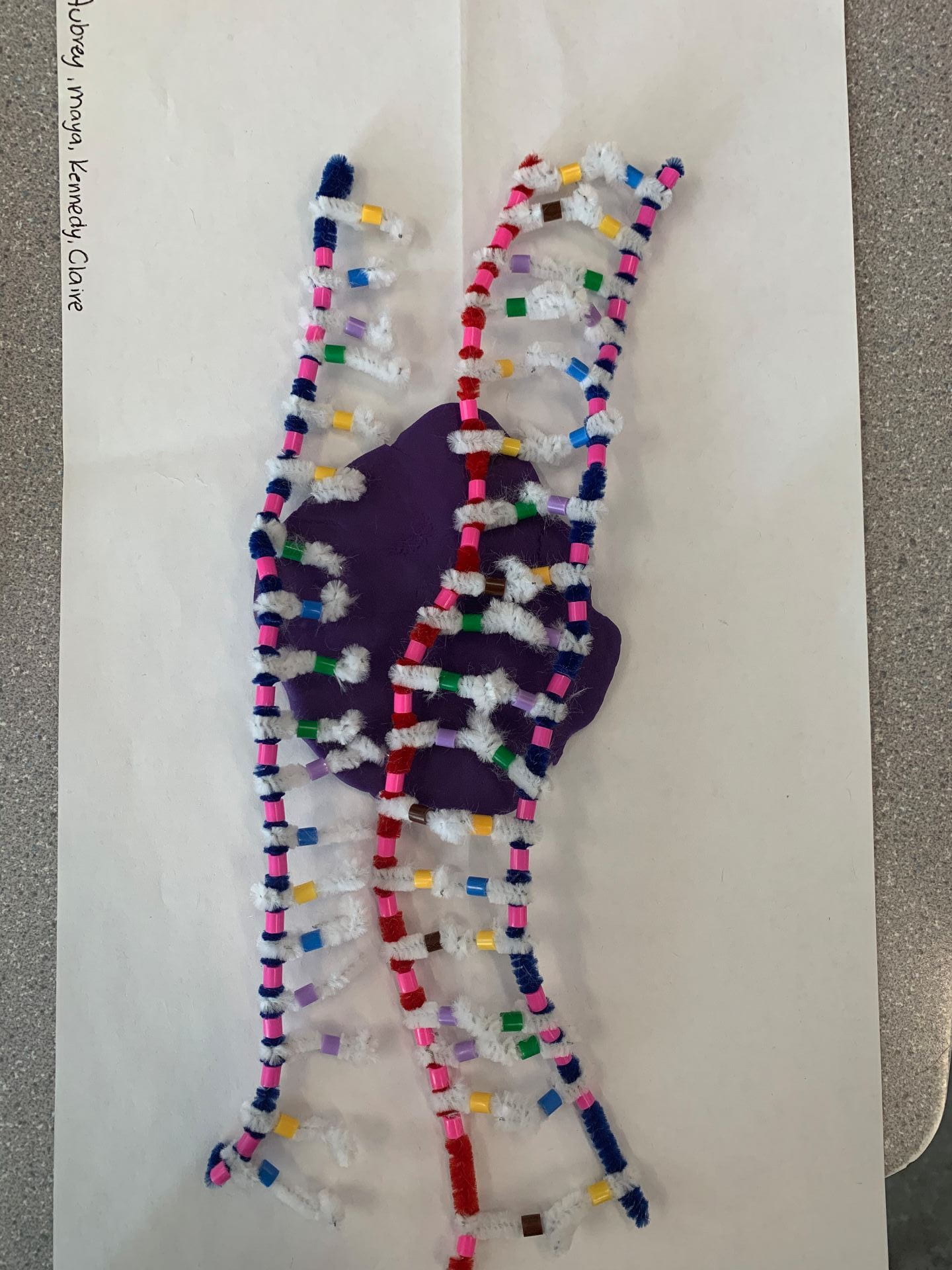 (our model was done incorrectly and does not accurately depict this stage of Transcription. GFSHRWHSHHRW
(our model was done incorrectly and does not accurately depict this stage of Transcription. GFSHRWHSHHRW
The final step in DNA Transcription is Separation from DNA. The mRNA removes itself from the DNA after the entire gene has been transcribed. The DNA returns to its double helix shape by zipping and winding back together. The mRNA is now leaving the nucleus and starting the translation process. At this step, we should be left with a DNA molecule is a double helix shape and an mRNA molecule on a mission with its message. To put it in very simple ways this is a complicated way for DNA to send a text message.  (This image similar to the previous one is also not completely correct. They are not in their correct shape and they should not be touching.)
(This image similar to the previous one is also not completely correct. They are not in their correct shape and they should not be touching.)
3. How does this activity do a good job of modeling the process of RNA transcription? In what ways was our model inaccurate?
This activity similar to the previous DNA model activity was helpful in modeling RNA transcription as it shows each individual step and really walks us through every part of DNA transcription. However, this activity is not 100% accurate as Transcription is supposed to be a small portion of the DNA strand however, our model shows the entire strand being transcribed. Furthermore, DNA is much larger than RNA in reality as RNA is approximately 1000 nucleotides and DNA is approximately 85 000 000 nucleotides. However, despite these inaccuracies, the activity still modeled Transcription very well.
4. Describe the process of translation: initiation, elongation, and termination.
Translation immediately after transcription. It is the process of translating the sequence of a messenger RNA (mRNA) into a sequence of amino acids.
The first step is Initiation. mRNA is being helped by the ribosome. The P site (The P site, called the peptidyl site, binds to the tRNA holding the growing polypeptide chain of amino acids) of the ribosome reads the AUG start Codon on the mRNA strand (the most common start spot is AUG.) The tRNA will bring in the first amino acid and then it will continue this process (elongation) until a stop codon (UGA) is reached.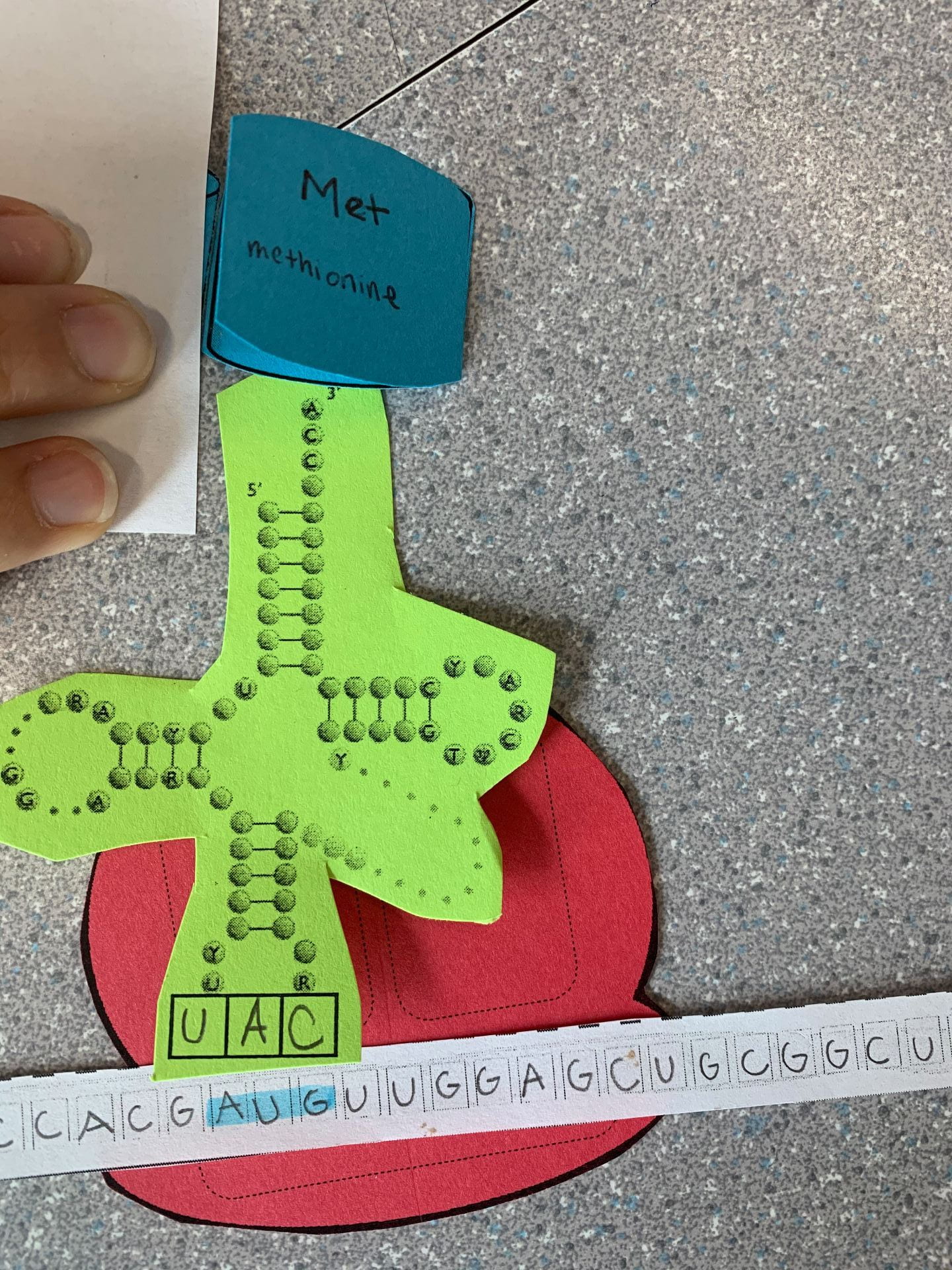
The second step is Translation is Elongation. In this step, the amino acid chain continues to grow hence the name Elongation. The A-site (The A site is the point of entry for the aminoacyl tRNA) reads the next mRNA codon and brings the matching tRNA. The amino acid chain from the tRNA in the P-site is transferred to the tRNA in the A-site. This continues to create a polypeptide chain.
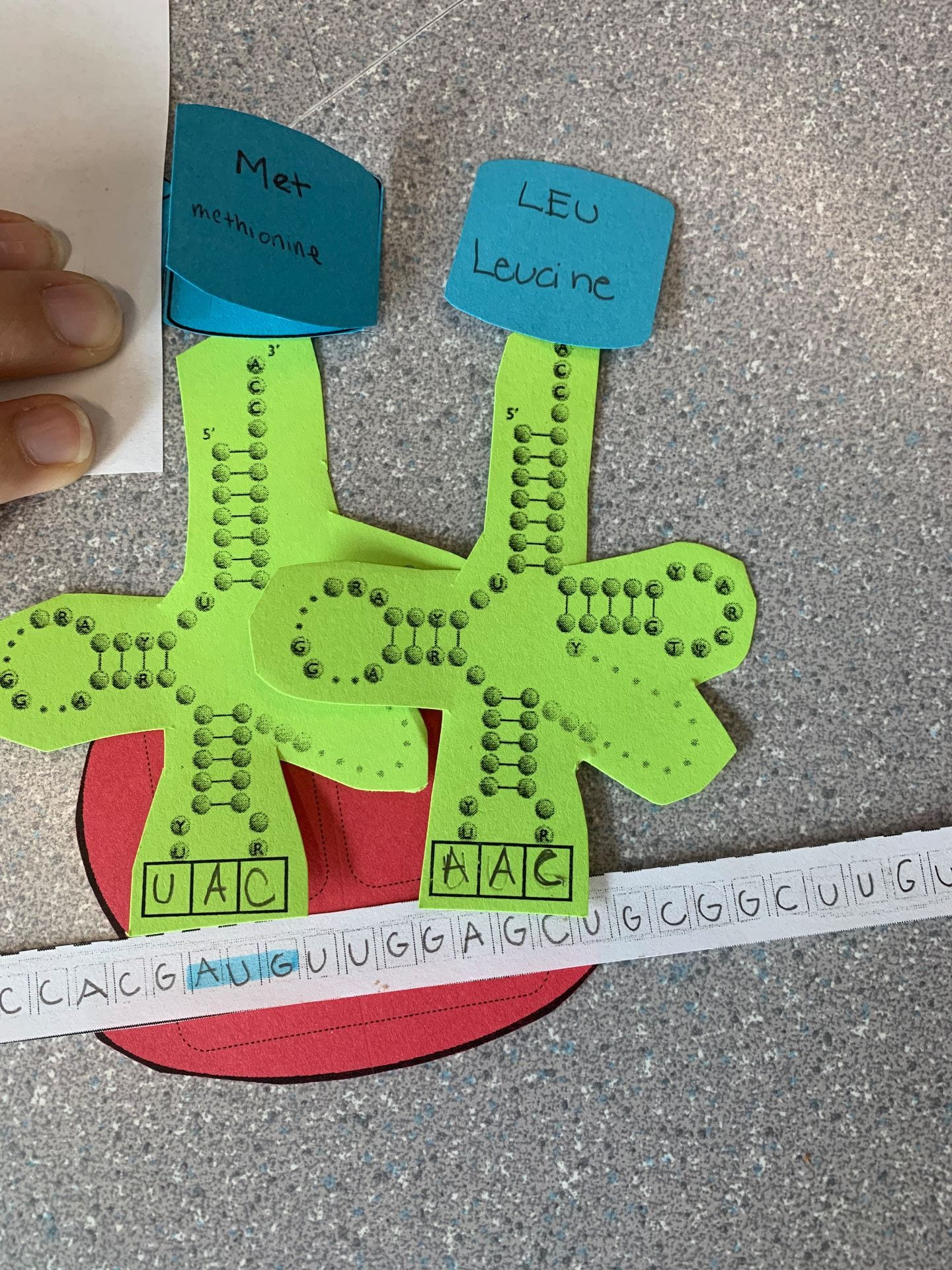
The final step in Translation is called Termination. This is the last stage of translation and allows for the polypeptide chain to become a fully functioning protein. In order for this to occur, we must reach one of the three stop codons. Stop codons are sequences of DNA and RNA that are needed to stop translation or the making of proteins by stringing amino acids together. There are three RNA stop codons: UAG, UAA, and UGA. There are no more amino acids onward. The ribosome lets go of the mRNA and the tRNA lets go of the polypeptide (Note that the blue pieces of paper should not be side by side and should be one above the other on top of the green pieces of paper from the previous image)
(Note that the blue pieces of paper should not be side by side and should be one above the other on top of the green pieces of paper from the previous image)
5. How does today’s activity do a good job of modeling the process of translation? In what ways was our model inaccurate?
Unfortunately, on the day of the activity, I was homesick and could not complete the activity in class. I may not be able to say that the in-class activity was helpful, however, the blog was helpful as I could retrace my group’s steps and get a better understanding of the process by using typing it out in my own explanation. As mentioned earlier I was not there for the activity, however, my group members filled me in on the ups and downs of the model and some mentioned inaccuracies is that the papers did not line up properly and there are some photos missing from our activity.





 ( these structures will be in the double helix form)
( these structures will be in the double helix form)
- Explain the structure of DNA.
Deoxyribonucleic acid also is also known as DNA is made up of monomers called nucleotides which are composed up of a 5-carbon sugar, a phosphate group and a nitrogenous base (Purines: Adenine, Guanine, and Pyramindines: Cytosine, and Thymine). DNA is made up of two antiparallel strands going in opposite directions but sending the same messages. This is represented in the photos by having one strand start with phosphate and one starts with sugar. The sugar is represented by a pink bead. Our model of the DNA has a minor mistake the Adenine and the Guanine should be represented by two beads as they are a purine with a double-ringed structure as opposed to the pyrimidines which have a single-ringed structure. The complementary base pairings are the nitrogenous bases that join together to form the DNA and a specific purine will always have a specific pyrimidine ( Adenine and Thymine together and Cytosine and Guanine together). These can be in mixed order, however, the A will always be with T and the C will always be with G in DNA.
2. How does this activity help model the structure of DNA? What changes could we make to improve the accuracy of this model?
This activity helps model the structure of DNA as we can clearly see all the important details that could be missed when just reading about DNA. This clearly demonstrates that DNA is antiparallel and one strand starts with sugar while the other starts with phosphate. Furthermore, this activity helps us memorize the base pairings and see the various steps of DNA as we create the model. Some changes to be made to help improve the accuracy of this model would be to bring things slightly more the scale. Of course, not it could not be exact as DNA is so incredibly small, however, ensuring the white pipe-cleaners are slightly more to scale to the beads and blue pipe-cleaners could help us visualize clearly what DNA looks like and functions in real life.
3. When does DNA Replication Occur?
DNA replication occurs immediately before cell division as it is needed for the cell to divide. DNA replication is necessary for all living organisms as it is what allows the genetic information to passed down and inherited to the next organism. DNA replication is known as a semi-conservative process as the replicated DNA contains part of the “old/original” DNA and also a “new/replicated” strand of DNA to create a new strand of DNA in a double-helix form.
4. Name and Describe the 3 Steps Involved in DNA Replication. Why Does the Process Occur Differently on the “Leading” and “Lagging” Strands?
Unwinding and Unzipping
- This is considered the first step in DNA replication. This is when DNA Helicase moves along the center of the strand and unravels the double helix structure to make more a ladder structure and moves along the complementary base pairings to break the bonds between them and separate the DNA completely. DNA helicase is an enzyme built to unzip the DNA in DNA replication.
Complementary Base Pairings
- The enzyme Polymerase uses the new complementary nucleotides (present in the nucleus) and pairs them with the original strand that was unzipped. DNA Polymerase can also only build from 5′ to 3′. It can start on the 3′ to 5′ original strand and work down, this is known as the “leading” strand. However, with the other original strand being 5′ to 3′, DNA Polymerase has to build in the reverse direction by making fragments since it only builds 5′ to 3′. This creates Okazaki fragments (Okazaki fragments are short sequences of DNA nucleotides that are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication). As DNA Polymerase works to build on this strand, there are constant interruptions along it making it known as the “lagging” strand.
Adjacent Nucleotides Joining
- This is known as the final step in DNA replication. In this step, the complementary base pairings are bonded together with the sugar-phosphate backbone to form a new DNA strand. The enzyme DNA ligase goes along the lagging strand to join together the DNA and repair the Okazaki fragments. The DNA then winds into a double-helix and this the replica of the DNA.
5. What Did You do to Model the Complimentary Base Pairing and Joining of Adjacent Nucleotide Steps of DNA Replication? In What Ways was This Activity Well Suited to Showing This Process? In What Ways Was it Inaccurate?
The way our group modeled each step of DNA replication was undoing the original strand of DNA and half hooking it to the “new/replicated” strand of DNA; therefore, showing that it is bonding to a new strand but not yet completed. To represent the stage of Adjacent Nucleotides steps with fully hooked together with the new strands of DNA to show the complete replication of DNA. This activity was well suited to show the complete process in each individual step as we needed to be able to assemble what each step looked like. However, this activity was inaccurate as you cannot model the Okazaki Fragments within the lagging strand of DNA and it cannot model the DNA Ligase moving along the lagging strand to repair any Okazaki Fragments.
 ( This is a photo of the agar cubes cut in half to see the diffusion after a 10 minute period.).
( This is a photo of the agar cubes cut in half to see the diffusion after a 10 minute period.).

 (our model was done incorrectly and does not accurately depict this stage of Transcription. GFSHRWHSHHRW
(our model was done incorrectly and does not accurately depict this stage of Transcription. GFSHRWHSHHRW (This image similar to the previous one is also not completely correct. They are not in their correct shape and they should not be touching.)
(This image similar to the previous one is also not completely correct. They are not in their correct shape and they should not be touching.)

 (Note that the blue pieces of paper should not be side by side and should be one above the other on top of the green pieces of paper from the previous image)
(Note that the blue pieces of paper should not be side by side and should be one above the other on top of the green pieces of paper from the previous image)



 ( these structures will be in the double helix form)
( these structures will be in the double helix form)