Explain the Structure of DNA
DNA or Deoxyribonucleic acid is made out of sugars (deoxyribose), phosphates, and nitrogen bases. DNA is a polymer that is made of nucleotide monomers. DNA has 2 sugar-phosphate backbones which form 2 antiparallel strands. These strands twist into a double helix shape. Between these strands or backbones we find the nucleotides: Adenine and Guanine (purines) and Cytosine and Thymine (pyrimidines). Each nucleotide has a complementary base. Adenine will always pair up or form an H-Bond with Thymine, and Guanine will always pair up with Cytosine.



How does this activity help model the structure of DNA? What changes could we make to improve the accuracy of this model?
This model emphasizes the complimentary base pairing. The different colours help to show where the phosphate is found in the backbone and the pattern of the complimentary base pairs. We are able to see how the purines and pyrimidines bond with each other. The white pipe cleaners do a good job at representing the H-Bonds between the nucleotide bases. One change that could be made to improve the accuracy of this model is to use specific measurements. To increase accuracy we would have the measurements of our pipe cleaners as well as the spacing between the pipe cleaners and beads have similar proportions of actual DNA. This would allow us to better understand the proportions between how long a DNA strand is compared to how wide it is.
When does DNA replication occur?
DNA replication occurs right before cell division. Before a cell can divide, it will need its own copy of instructions or DNA to keep in its nucleus.
Name and describe the 3 steps involved in DNA replication. Why does the process occur different on the “leading” and “lagging” strands?
Step 1: Unwinding and Unzipping
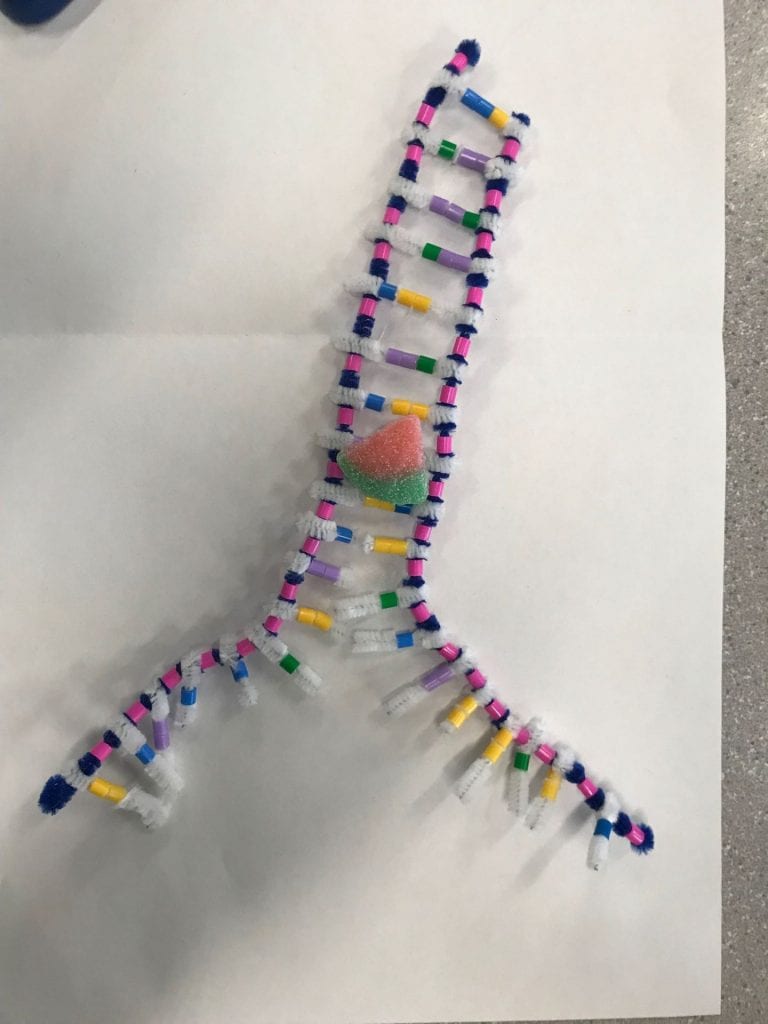
Step 2: Complimentary Base Paring
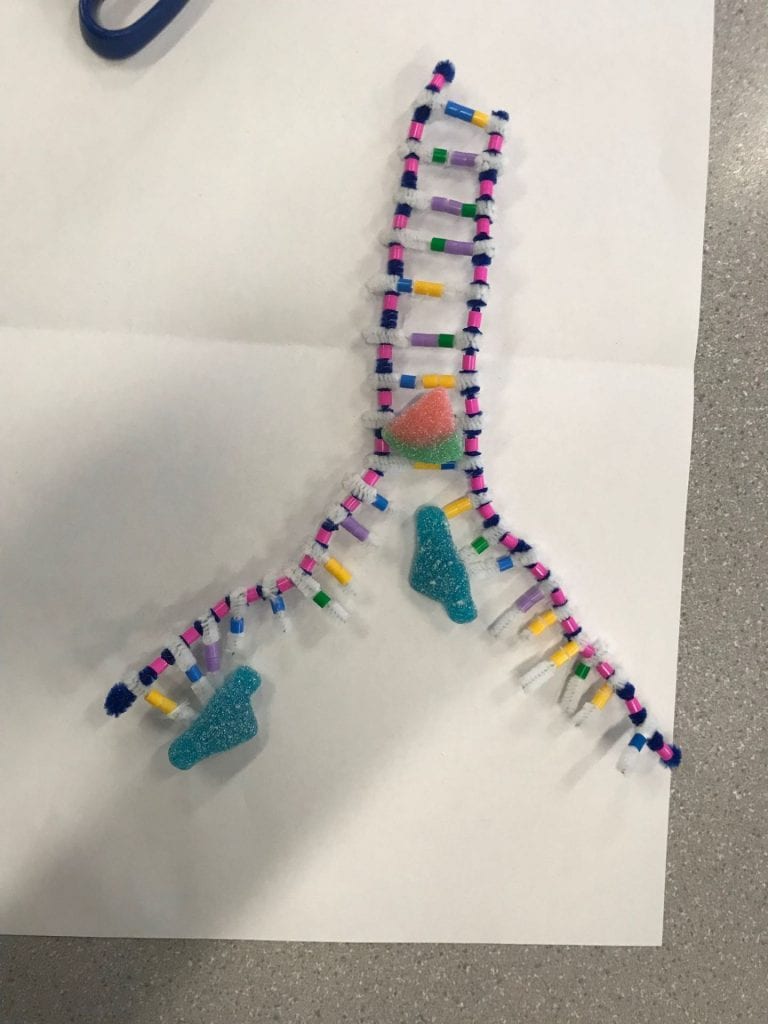
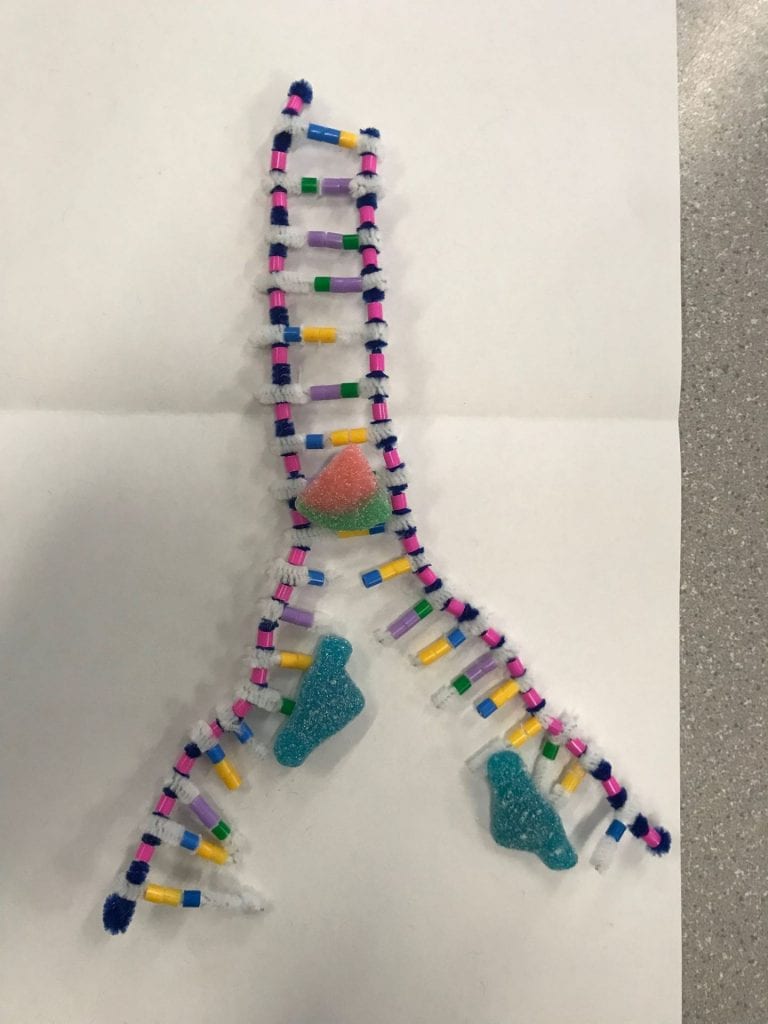
Step 3: Joining adjacent nucleotides

In step 2, we stated that during complimentary base pairing, the DNA polymerase has to follow a certain synthesis direction. It must travel away from the phosphate end and towards the sugar end. The strands are antiparallel, meaning the synthesis direction is different for each strand which results in a “leading” strand and a “lagging” strand. The nucleotides are added differently to the leading and lagging strands. The DNA polymerase on the leading strand simply follows the helicase. The lagging strand, on the other hand, goes in the opposite direction of the helicase. As the helicase unwinds and unzips more of the DNA, the polymerase of the lagging strand has to go back in order to complete the complimentary base pairing. In summary, the polymerase of the leading strand will add the nucleotide bases all in one segment. The polymerase of the lagging strand will have to complete the complementary base pairing in multiple segments. These segments will later be attached together in the 3rd step by the ligase.

What did you do to model the complimentary base paring and joining of adjacent nucleotides steps of DNA replication. In what ways was this activity well suited to showing this process? In what ways was it inaccurate?
To model the steps of the complimentary base pairing we first separated a section of the DNA molecule. Once we had the strands partially separated, we then added the appropriate base pairs. This activity helped break down the 3 different steps and provide a better understanding of the responsibility of each enzyme. We could clearly see and understand that the helicase was in charge of unwinding the DNA from it’s double helix form and separating the strands. This activity was able to provide a basic demonstration on the difference between the lagging and leading strands as well. As we created the two diagrams for step 2, we followed the basic directions that the polymerase would’ve followed. When we were adding the complimentary bases onto the leading strand, we followed the strand in the same direction throughout the entire process. As more of the strand was separated, we continued to travel upwards. On the lagging strand, you would add nucleotides until you reached the end of the strand. Once more of the strand was separated you would have to back up, past your initial staring place, and synthesize downwards until you met up with your original segment.
This activity is quite inaccurate in showing the actual movements of the enzymes. This activity isolates the enzymes so we don’t see how these enzymes would be working on this DNA molecule at the same time. In the activity process, we move each enzyme individually, however in real life, the enzymes would be working simultaneously. I also found this activity was inaccurate at modelling the process of the ligase enzyme. The photo is decent but the way you would manipulate your model to create that photo is not reflective of the real life process.
RNA Transcription
How is mRNA different than DNA?

Both mRNA and DNA have sugar-phosphate backbones but the sugars used for each of them are different. Like their names indicate, DNA uses deoxyribose while RNA uses ribose. Another key difference between the two is that mRNA is able to leave the nucleus while DNA cannot. mRNA or messenger RNA serves to deliver DNA’s messages to places outside of the nucleus.
Describe the process of transcription



How did today’s activity do a good job of modelling the process of RNA transcription? In what ways was our model inaccurate?
Today’s activity helped the break down the steps of RNA transcription. The activity demonstrates how the DNA behaves and how it is used as a template in the formation of mRNA. This activity was also a great way to compare the similarities and differences between the replication of DNA vs the transcription of RNA. This model only shows the gist of what happens during each step. Not all the processes and enzymes that would be used are shown. For example, the description of RNA polymerase is quite basic. The task of the polymerase is clear but how it goes about doing that is not as clear. This activity did not show how mRNA is modified in preparation of leaving the nucleus as well. This activity also inaccurately shows mRNA as being the exact same length as the DNA strand.
Protein Synthesis
RNA Transcription


Describe the process of translation
- Initiation


2. Elongation


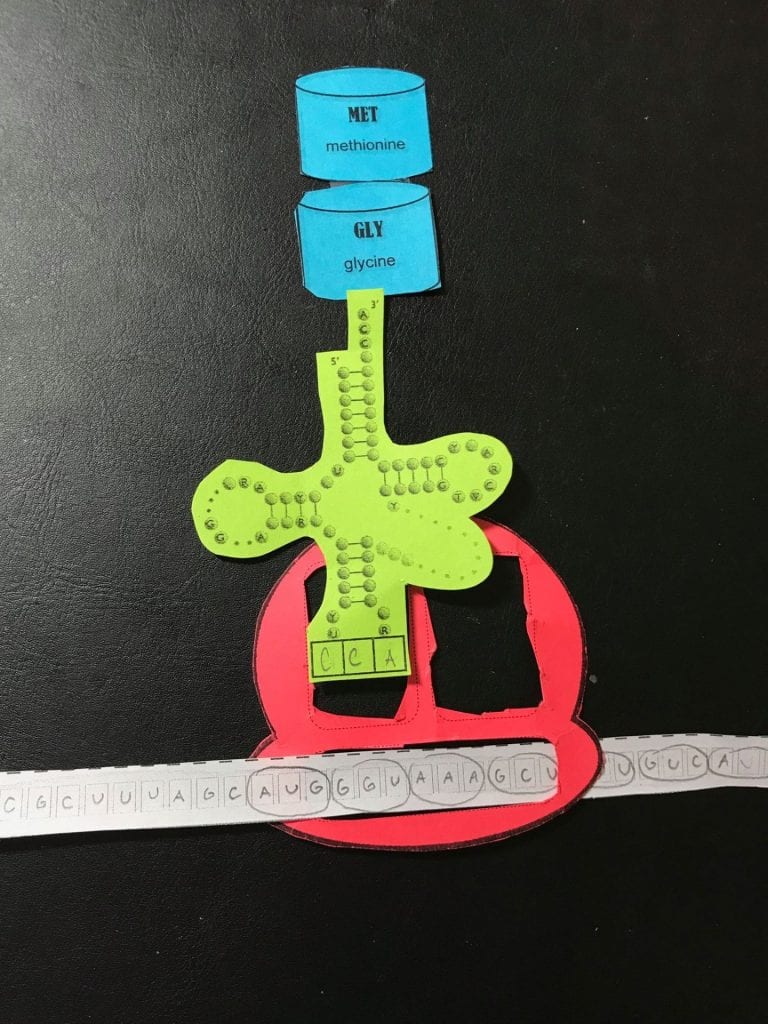


3. Termination


How did today’s activity do a good job of modelling the process of translation? In what ways was our model inaccurate?
Today’s activity did a great job at accurately breaking down and modeling the steps of translation. The paper cut outs were very clear and allowed for the activity to be quite hands on. This activity really helped me to understand the movements and actions that occur during elongation. The cut outs allowed me to be hands on, for example, I would physically move the tRNA or the amino acids from the different sites. One thing that was inaccurate with this model is that we showed that only one ribosome was translating this mRNA. In real life, there would be multiple ribosomes that are simultaneously going along the mRNA and making the protein.

