By: Keisha N, Yinling C
Purpose: To determine the effects of temperature on the reaction rate of the enzymes reactions
Hypothesis: The high temperature will cause a higher reaction rate causing more glucose to appear.
When heated above 45 degrees the protein will denature and there will be no glucose. The lower temperature will cause a slower reaction rate causing less glucose to appear.
Materials:
- Lactase enzyme drops
- (lactose) Milk (100mL)
- 1L of water
- 5 Glucose test strips
- 5 Test tubes
- Test tube rack
- 1 stir stick
- Thermometer
- Bowl of ice (ice bath)
- Heat source (hot water bath)
- Three beakers
Procedure:
- Fill one beaker with 500mL of water and place on the hotplate (medium – high heat)
- Fill the second beaker with 500mL of ice water (ice water bath)
- Fill each of the test tubes with 20mL of the lactose milk
- Add 2 drops of lactase enzyme drops to each of the test tubes
- Take one test tube and place it in the test tube rack to act as the control group, Make sure it is at room temp (23ºC). Keep for 10 minutes.
- Take one test tube and place in the ice bath once it has dropped to 15ºC. Place in the bath for 10 minutes making sure that the temperature stays within close range.
- Take the third test tube and place it in the hot bath once it has reached 28ºC, leave the test tube in the hot bath for 10 minutes. Check temperature range regularly.
- Once 10 minutes has passed, take the test tubes out of the baths. Place a sample from each test tube on the test strips. Record colour. Repeat for beakers with body temperature and for 45ºC.
- Take the fourth test tube and place it in the hot bath, once it has reached body temperature (37ºC), leave the test tube in the hot bath for 10 minutes.
- Take the last test tube and place it in the hot bath once it has reached 45ºC, leave the test tube and place in the bath for 10 minutes.
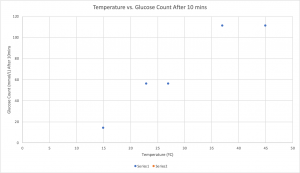
Data and Observations:
| Beaker | Temperature
|
Colour of the Test Strips | Presence of Glucose (mmol/L)
|
| 1 | 15ºC | Green | 14 mmol/L |
| 2 | Room Temperature (23ºC) | brown
Olive Green |
56 mmol/L |
| 3 | 28ºC
|
Light Brown | 56 mmol/L |
| 4 | 37ºC | Light Dark Brown | 111 or more mmol/L |
| 5
|
45ºC | Brown | 111 or more mmol/L |
Questions:
1. At what temperature was there the lowest count of glucose? – Why?
The lowest amount of glucose was present at 15ºC. This was because when you lower the temperature of the milk it causes the reaction rate of the enzymes to slow down, causing there to be less glucose present.
2. What would happen to the enzymes if it were placed into a test tube with a temperature higher than 45ºC?
If placed in a test tube above 45ºC there should be no glucose present in the milk. This is due to the fact that when an enzyme is heated to above 45ºC, the enzyme will start to denature, causing there to be no trace of glucose.
3. Why does 37ºC (body Temperature) produce the most optimal conditions for enzymes to work in?
As the temperature rises, reacting molecules have more kinetic energy; this increases the chances of successful collisions and thus, the reaction rate increases. At 37ºC, the enzyme’s catalytic activity is at its greatest and therefore making it the most optimal temperature for the human cell.
Conclusion:
This lab allowed us to determine the effects of temperature on the reaction rate of the enzymes. We had hypothesized that there would be little to no glucose present when the temperature of the milk was lowered and that there would be high amounts of glucose when the temperature was raised, our hypothesis proved to be correct. We also predicted that no glucose would be present at 45ºC which was proven wrong. The incorrect hypothesis could be due to an error during the lab. At the lowest temperature (15ºC) there was little to no glucose present due to the low temperature and slow reaction rate. At he high temperatures we found that like we had hypothesized we found that there were high amounts of glucose present due to the high temp. and the increased reaction rate. When we had heated up the milk to 45ºC, the enzyme was supposed to denature however, due to error this was not the case. The most glucose was present at 37ºC and 45ºC. This lab gave us the ability to learn more about the relationship between temperature and how it effects enzymes.
Errors/Improvements:
To improve our lab design, we would have kept the amount of tests that we did however change the temperatures at which it happened. We tested too many temperatures that were in the same range and not much of a variety. We should have done at least another test at a very low temperature and one above 45ºC. It was also difficult for us to maintain a constant temperature causing there to be an error in our lab. The enzyme should have denatured once heated to 45ºC or higher.