Explain the structure of DNA
DNA also known as deoxyribonucleic acid is a macromolecule made up of monomers known as nucleotides which include a phosphate group, a nitrogenous base and 5-carbon sugars. There are 2 types of nitrogenous bases purines and pyrimidines. Cytosine and Guanine are pyrimidines and single ringed structures where Adenine and Guanine are purines and double-ringed structures.
DNA is composed of two strands that are antiparallel, they run in opposite directions but still relay the same message. One strand runs 5′ to 3′ and the other strand runs 3′ to 5′. This just shows the position of the phosphates and sugars on the backbone.
The white pipe cleaners in the images below represent the hydrogen bonding occurring between the two strands called complementary base pairing. Complementary base pairing is when Adenine and Thymine only bond with one another because they form with 2 hydrogen bonds and Guanine and Cytosine only bond with each other because they bond with 3 hydrogen bonds.
After the two antiparallel strands are bonded together through complementary base pairing (shown in the 2nd image) the strand twists due to the force of the bonds to form into a double helix shape (shown in the 3rd image).



How does this activity help model the structure of DNA? what changes could we make to improve the accuracy of this model?
The pipe cleaner activity was a good activity to understand the structure of DNA and help visualize its shape and structure. The images we see online don’t give as good of an idea of the actual shape of DNA. Having an actual model and creating it ourselves really helped me to understand certain parts I didn’t notice in class from lessons and pictures. One thing that we could change to make this activity more accurate would be to show how many bonds are between each nitrogenous base (either 2 or 3).
DNA Replication
When does DNA replication occur?
DNA replication occurs in all living organisms before cell division. It is necessary for genetic information to be passed down and inherited. DNA replication is known as a semi-conservative process where a replicated strand will contain an original or old strand as well as have a new or replicated strand to make a complete new DNA strand.
Name and describe the 3 steps involved in DNA replication. Why does the process occur differently on the leading and lagging strands?
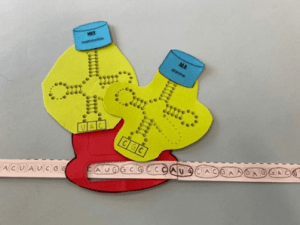
- Unwinding and Unzipping: Unwinding and unzipping occurs when DNA helicase unwinds the double helix and works to break the hydrogen bonds between the nitrogenous bases. Shown in the image below
- Complementary Base Pairing: DNA polymerase uses individual nucleotides floating within the nucleoplasm to pair with the original strands that were unzipped. It only builds in a 5′ to 3′ direction therefore it can start on the 3′ to 5′ original strand and work down without interruption, it is known as the leading strand. The second original strands is 5′ to 3′ and therefore has to build backwards creating Okazaki fragments. This strand is known as the lagging strand as there are constant interruptions
- Adjacent Nucleotides Joining: The last step is when the nucleotides that were bonded to their complementary bases are bonded together with the sugar-phosphate backbone. DNA ligase is responsible for the joining that ends in DNA winding into a double helix and finishing as a complete replica of the original strand.

1. unwinding and unzipping

2. complimentary base pairing

3. adjacent nucleotides joining

final image of the replicated strands
What did you think of the complementary base pairing and joining of adjacent nucleotides steps of DNA replication? In what ways was this activity well suited to showing this process? In what ways was it inaccurate?
To model the complementary base pairing and joining of adjacent nucleotides steps of DNA replication, we used 2 new strands of blue pipe cleaners which represented the sugar-phosphate backbones, with the replicated opposite side of bases with the beads, making it possible for unzipping strands to find a new strand to hydrogen bond with. This activity was good at displaying how the molecule unzips with the help of the enzyme polymerase. Our model shows how one molecule turns into two completely separate copy because of this unwinding process.
The activity was inaccurate because of the way it showed the hydrogen bonding and exactly where the original sugar-phosphate backbone ended up and how a new one forms in a real DNA replication. As well as the true double helix form wasn’t possible to create because of the materials we were using and they would hold the shape. Finally, the model doesn’t show how a parent DNA strand stays intact during replication.