DNA Structure
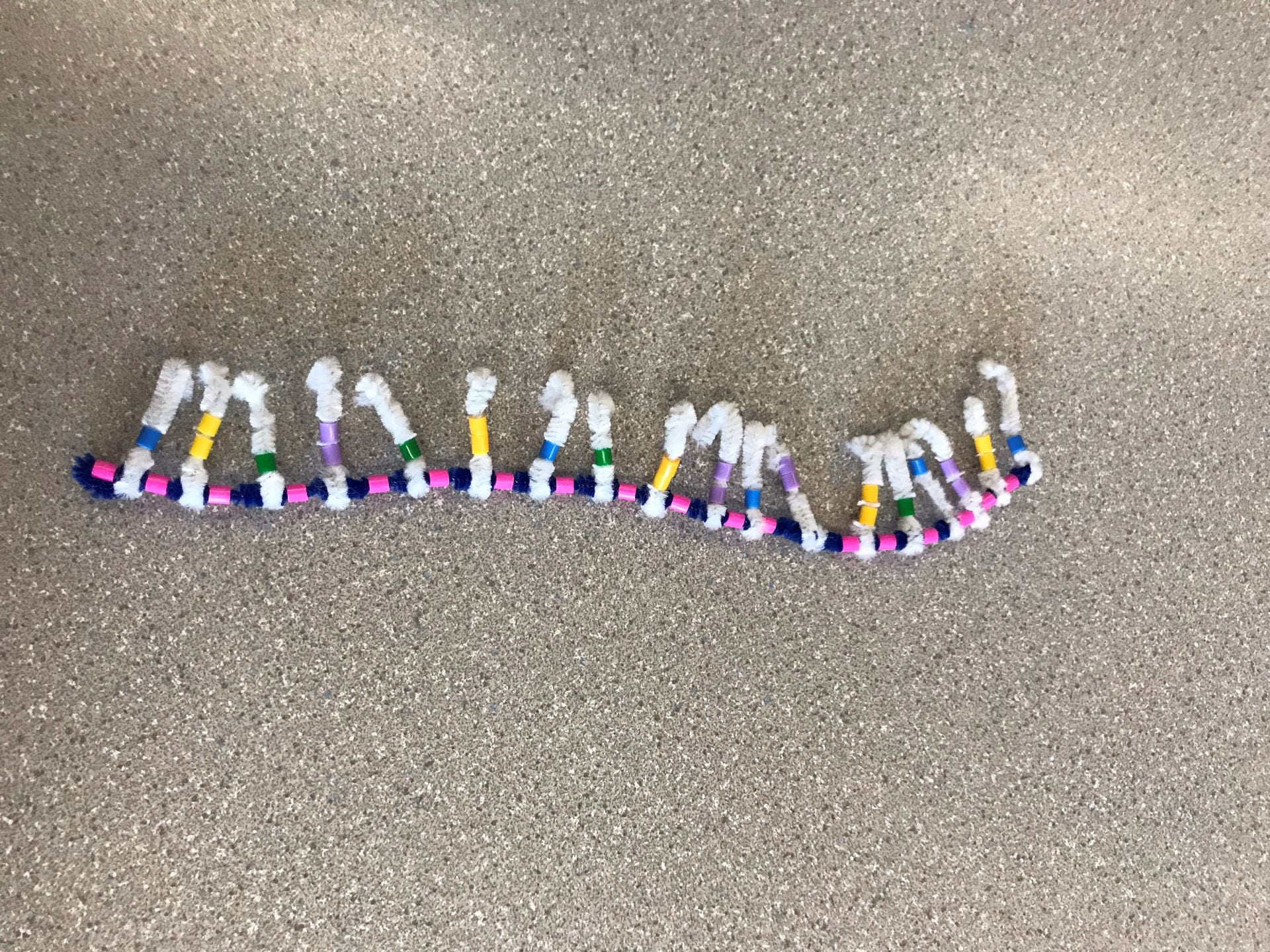
DNA is created by having two backbones of sugar-phosphate groups, as we can see with the blue pipe cleaner (the Deoxyribose sugar) and the pink beads (the phosphate groups). The order must always go in an alternating order of sugar-phosphate or vice versa. Each sugar-phosphate group has a nitrogen-containing group sticking out. The group of sugar-phosphate-base is the basic monomer for DNA which is called a nucleotide. Many nucleotides are bonded together to form a strand of DNA.
There are four different bases: Adenine (Yellow bead), Guanine (Purple bead), Cytosine (Green bead), Thymine (Blue bead). Adenine and Guanine are purines, which mean they have two rings. This is represented by Adenine and Guanine having two beads. Cytosine and Thymine are pyrimidines, which mean they have one ring. This is represented by Cytosine and Thymine having one bead. The bases follow a “protocol” called complimentary base pairing, which means that a certain base will only bond with another certain base. Adenine (A) will only bond with Thymine (T), and Guanine (G) will only bond with Cytosine (C).
The first strand will start with three specific bases, and this is the same for all strands of DNA. The sequence goes T, A, C. This means that the opposite strand will begin with the sequence A, T, G. From there, the sequence will differ for different strands of DNA, and there will be complimentary base pairing for the opposite strand.

When the strand is completed, the opposite, complimentary strand will be created following two different “rules”. The first is that the sugar-phosphate groups will be facing the opposite way. This means that if the first, original strand had a sequence of sugar-phosphate, the opposite strand will have a sequence of phosphate-sugar. This is called an antiparallel strand. The second is that the bases will be complimentary to the first strand following the complimentary base pairing rule. This means that the complimentary strand will begin with the three bases A, T, G. The bases in the two strands will H-bond with one another, creating a form that looks similar to a ladder.

Now that the ladder form is complete, the strand must twist into a double helix. DNA molecules have a right-handed twist and is twisted evenly throughout. The form of DNA that most people know is now complete.
The model of DNA and its characteristics are very well demonstrated by this activity. The diagram is almost identical to what the textbook models suggest. In the textbook diagrams, it is hard to see that the backbone is made of a sugar-phosphate group since it just looks like a backbone strand. The activity could be more accurate by putting the bases closer to one another, farther from the sugar-phosphate backbone. The last suggestion for this activity would be twisting the strand into a more accurate double helix. The pipe cleaners and the beads were prone to falling apart if they were twisted too much. If there was a certain way to allow the strand to twist more without breaking, that would be better.
DNA Replication
Cells, much like ourselves, age and become defunct. In that case, the cells must reproduce via cell division. However, the DNA in the parent cell nucleus must also be copied into the new cell nucleus. This happens via DNA replication. DNA must be copied and remade into a new strand of the same DNA before any cell division can occur. Therefore, DNA replication occurs when cell division happens.

There are three steps during DNA replication.
The first step is called the unwinding, or the unzipping process. In this step, the double helix untwists itself back into the ladder form and is unwounded/unzipped. This unzipping is facilitated by an enzyme called DNA Helicase, which breaks the H-bonds between nitrogen-containing bases so that the ladder is separated into the two separate backbones.

The second step is called Complimentary Base Pairing. This is the process where the unzipped strands begin to find their new “partner” strands. Nucleotides of all types are floating around in the nucleus. Complimentary bases to the original strand are brought in and connected into fragments. This process is facilitated by the enzyme DNA Polymerase. The direction in which DNA Polymerase reads and strand and brings in complimentary bases is strictly one-way; from Carbon-3 to Carbon-5 to be exact. For the leading strand, the sequence of the backbone molecules goes 5 to 3, which means that it follows a phosphate-sugar sequence. This means that as the DNA unravels, the DNA Polymerase can continue to read the strand and bring in complimentary bases. For the lagging strand, it is the opposite. The sequence of the backbone molecules goes 3 to 5 (sugar-phosphate sequence). This means that as the DNA unravels, the DNA Polymerase reads the strand in the same direction as the strand, which means that the DNA Polymerase has to return to the beginning of the unravelling Replication Fork and read again.

The final step is called joining. This is the process where fragments of complimentary bases are “glued” together into a long backbone strand. This is facilitated by the enzyme DNA Ligase. From there, once the long strand of complimentary bases are completed, they form H-bonds with their complimentary bases of the original strand. Now, the original strand has successfully formed two new DNA strands with two separate backbones. They will twist again and form a double helix.

It was difficult to show the individual nucleotides floating towards the replication site since we had not thought of cutting up the blue pipe cleaner to demonstrate this. Due to this, we had modeled the process of Complimentary Base Pairing and Joining virtually together. Furthermore, we did not have the best idea as to where the DNA Ligase should have been, so after looking at various diagrams on Google, we decided to simply put the Ligase near the fragment of complimentary bases. This would have been an inaccurate way of showing this model. Also, for the unravelling of the DNA, we were not entirely sure if the TAC sequence of the leading strand had to be the first to unravel, or the last to unravel. We ended up guessing that it would be the beginning that unravels first, and we’re still not sure whether that is the case or not.
However, there were many accurate representations in the model that we made. For example, the DNA Helicase looked mostly like the pictures of DNA Helicase that we saw in the notes or in textbooks. The general model looked like the diagrams in the notes as well. The way that we had shown the H-bond between the bases (hooking the white pipe cleaners together) made it relatively simple for us to unhook the “bond” and separate them to show the process of Complimentary Base Pairing.